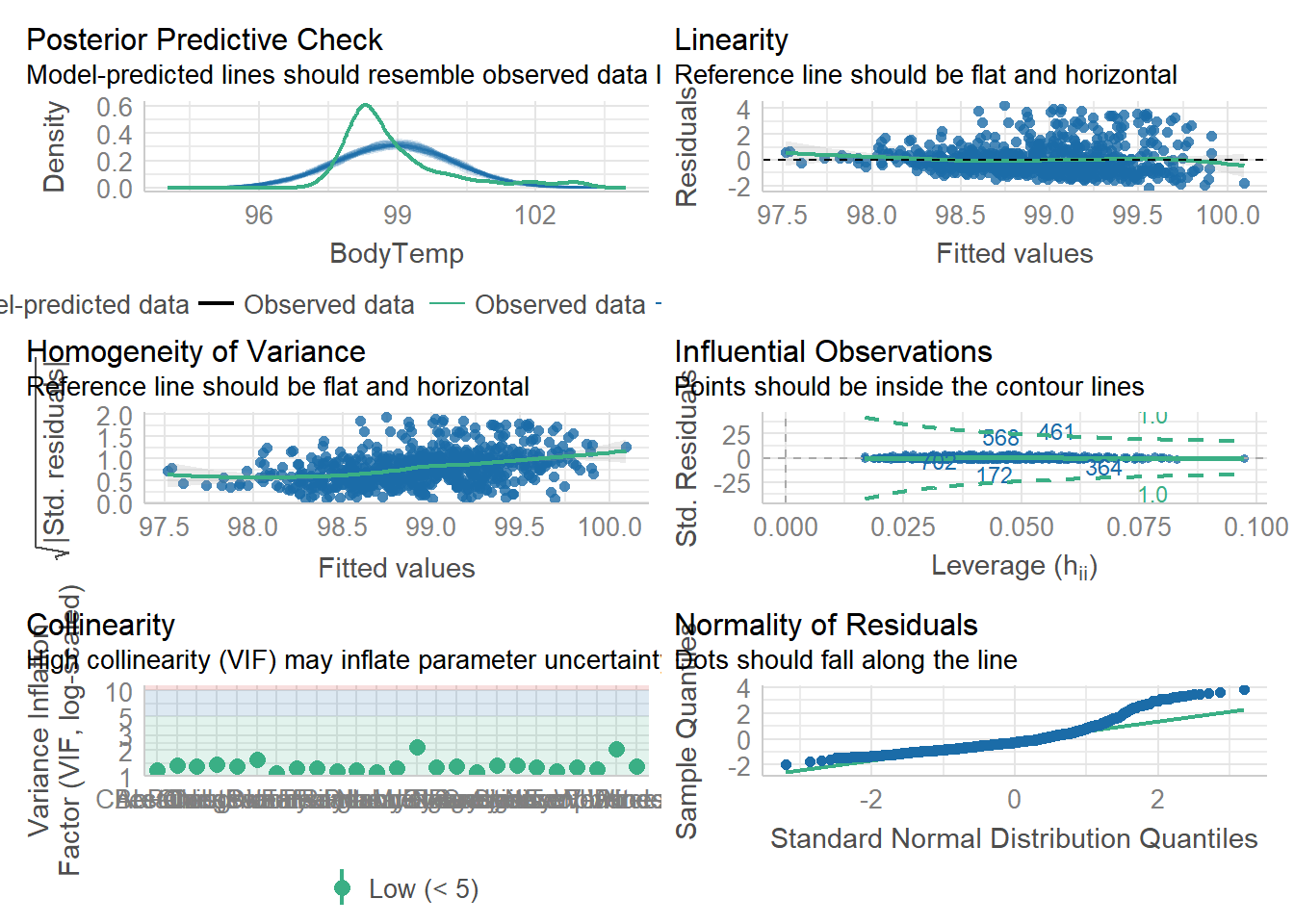
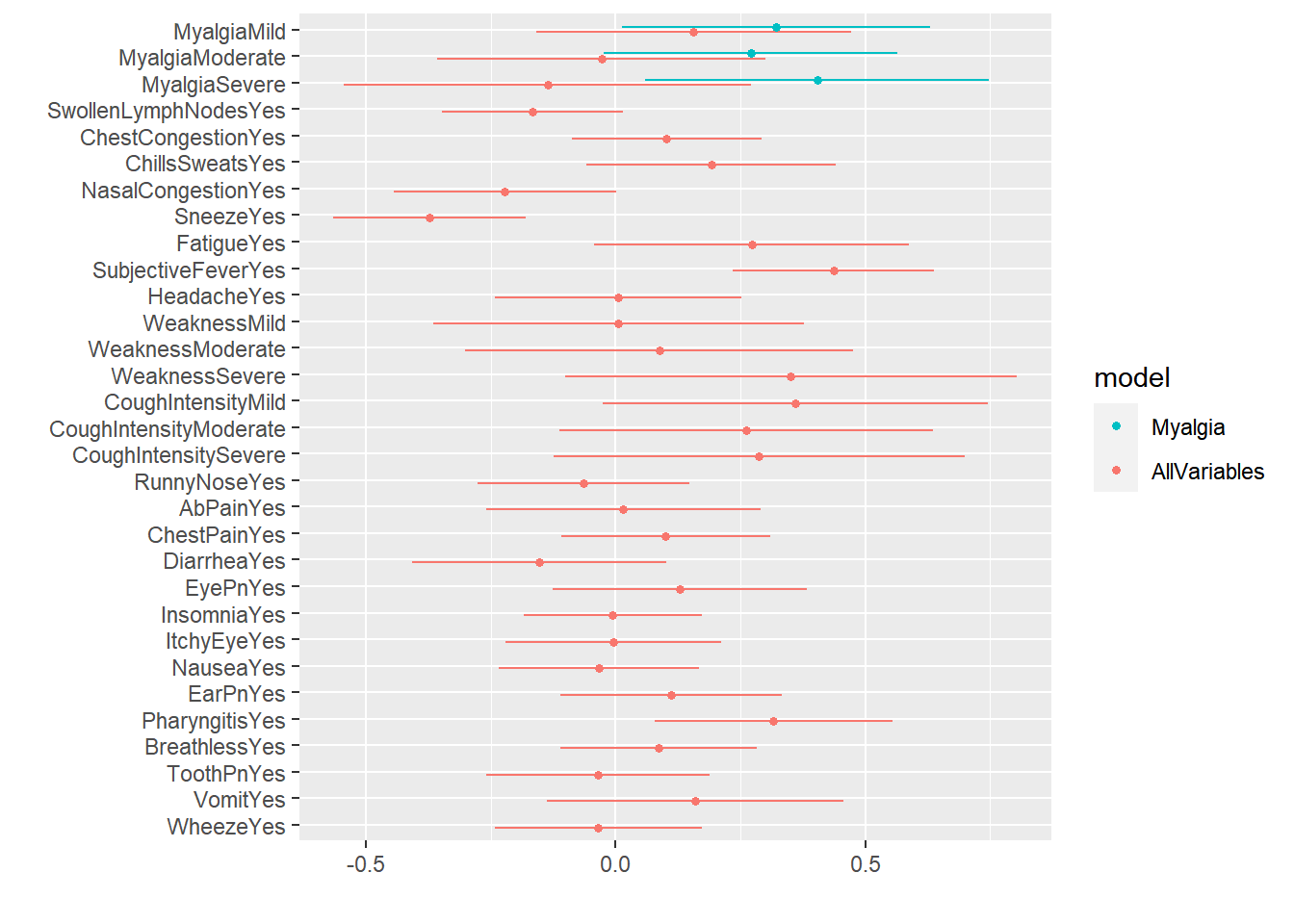
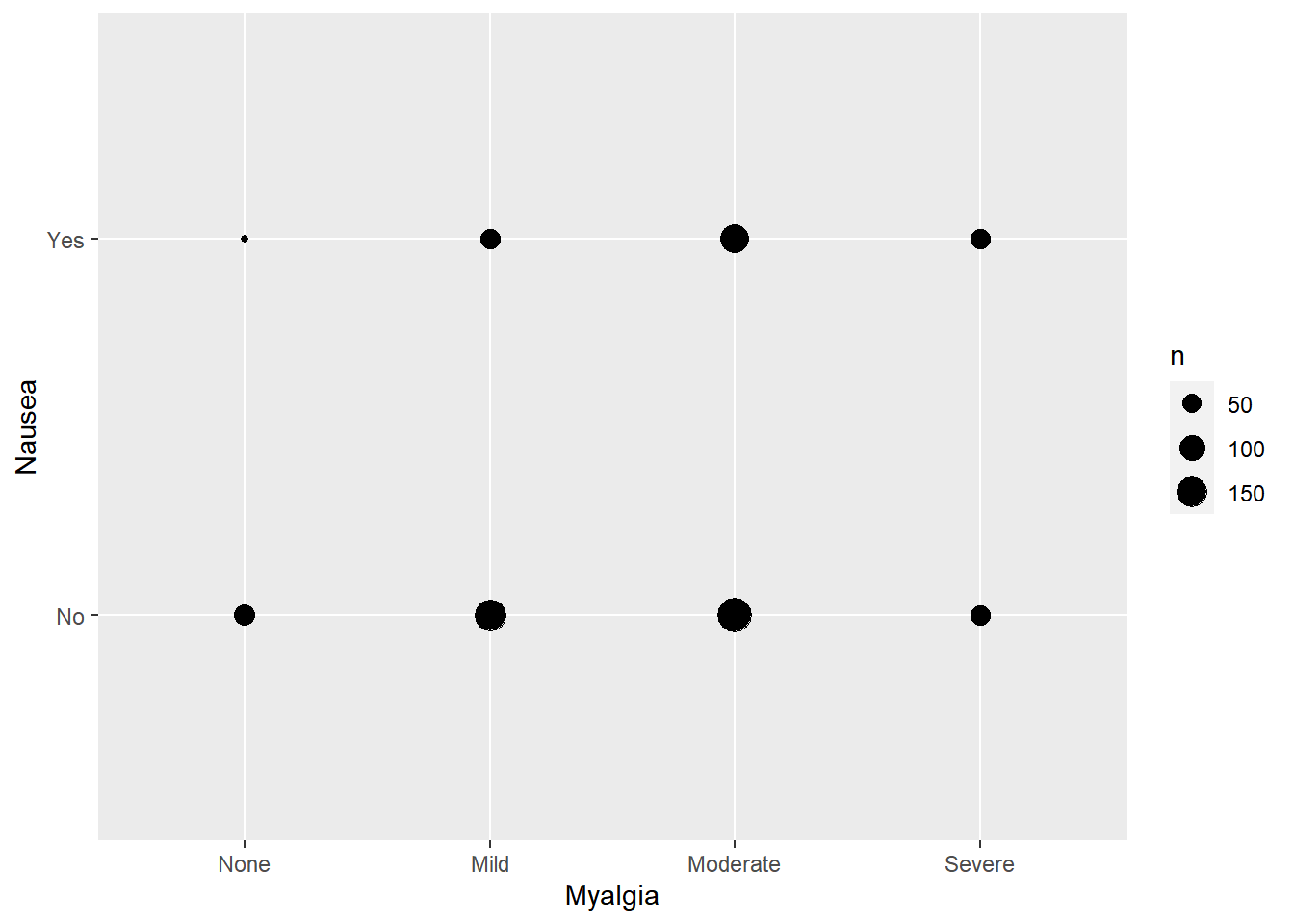
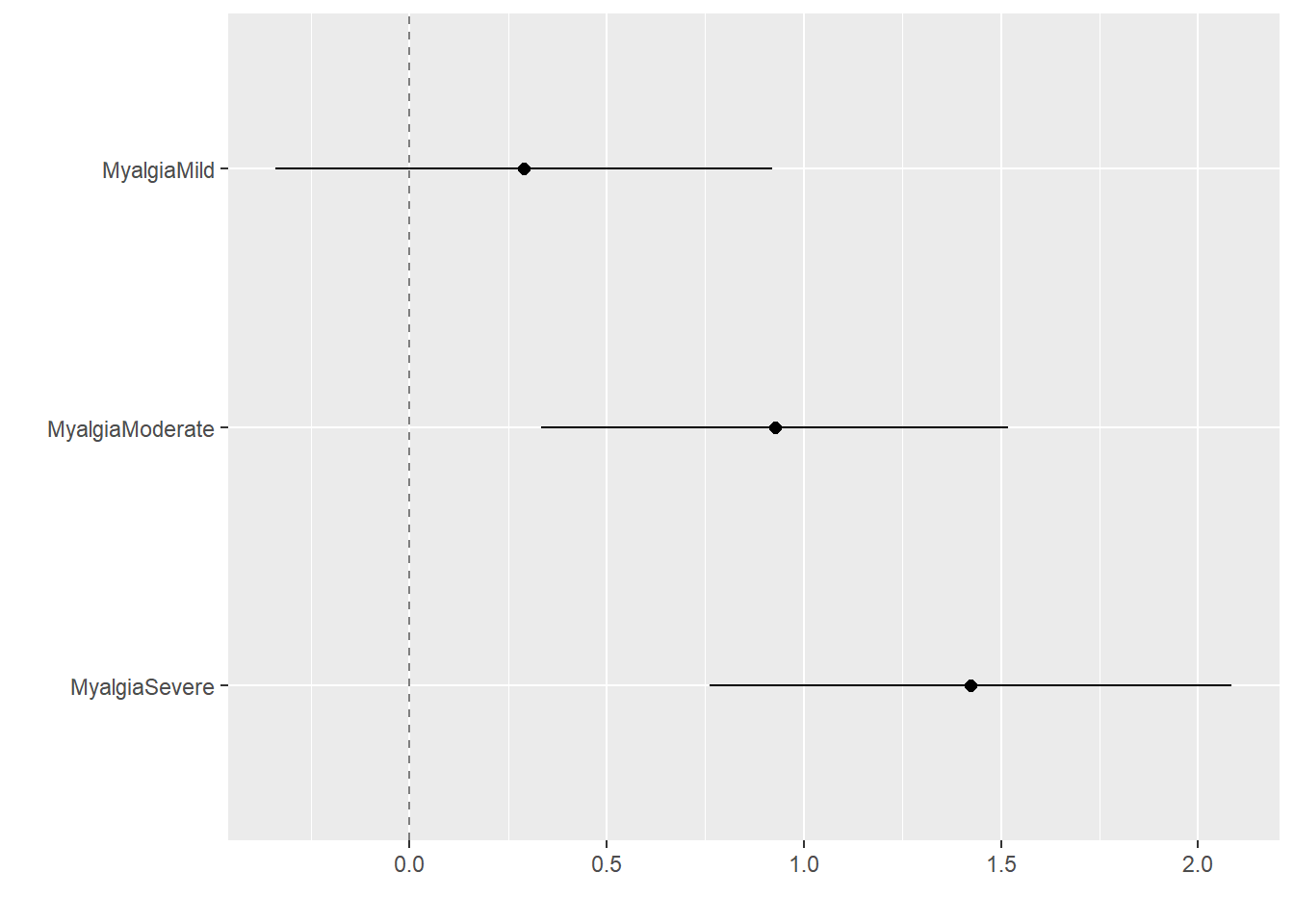
Warning: package 'tidyverse' was built under R version 4.2.2
── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
✔ ggplot2 3.4.0 ✔ purrr 1.0.1
✔ tibble 3.1.8 ✔ dplyr 1.0.10
✔ tidyr 1.2.1 ✔ stringr 1.5.0
✔ readr 2.1.3 ✔ forcats 0.5.2
Warning: package 'ggplot2' was built under R version 4.2.2
Warning: package 'stringr' was built under R version 4.2.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
library (tidymodels) #build models
Warning: package 'tidymodels' was built under R version 4.2.2
── Attaching packages ────────────────────────────────────── tidymodels 1.0.0 ──
✔ broom 1.0.3 ✔ rsample 1.1.1
✔ dials 1.1.0 ✔ tune 1.0.1
✔ infer 1.0.4 ✔ workflows 1.1.2
✔ modeldata 1.1.0 ✔ workflowsets 1.0.0
✔ parsnip 1.0.3 ✔ yardstick 1.1.0
✔ recipes 1.0.4
Warning: package 'broom' was built under R version 4.2.2
Warning: package 'dials' was built under R version 4.2.2
Warning: package 'infer' was built under R version 4.2.2
Warning: package 'modeldata' was built under R version 4.2.2
Warning: package 'parsnip' was built under R version 4.2.2
Warning: package 'recipes' was built under R version 4.2.2
Warning: package 'rsample' was built under R version 4.2.2
Warning: package 'tune' was built under R version 4.2.2
Warning: package 'workflows' was built under R version 4.2.2
Warning: package 'workflowsets' was built under R version 4.2.2
Warning: package 'yardstick' was built under R version 4.2.2
── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
✖ scales::discard() masks purrr::discard()
✖ dplyr::filter() masks stats::filter()
✖ recipes::fixed() masks stringr::fixed()
✖ dplyr::lag() masks stats::lag()
✖ yardstick::spec() masks readr::spec()
✖ recipes::step() masks stats::step()
• Use suppressPackageStartupMessages() to eliminate package startup messages
library (here) #help read/import data
Warning: package 'here' was built under R version 4.2.2
here() starts at C:/Users/Raquel/GitHub/MADA/RaquelFrancisco-MADA-portfolio
library (ggplot2) #data visualization library (broom.mixed) # for converting bayesian models to tidy tibbles
Warning: package 'broom.mixed' was built under R version 4.2.2
library (dotwhisker) # for visualizing regression results
Warning: package 'dotwhisker' was built under R version 4.2.2
library (performance) #evaluate model fit and performance
Warning: package 'performance' was built under R version 4.2.2
Attaching package: 'performance'
The following objects are masked from 'package:yardstick':
mae, rmse
<- readRDS (here ('fluanalysis/data/SypAct_clean.rds' )) #upload cleaned data tibble (data) # to get a look at the data
# A tibble: 730 × 26
SwollenLymph…¹ Chest…² Chill…³ Nasal…⁴ Sneeze Fatigue Subje…⁵ Heada…⁶ Weakn…⁷
<fct> <fct> <fct> <fct> <fct> <fct> <fct> <fct> <fct>
1 Yes No No No No Yes Yes Yes Mild
2 Yes Yes No Yes No Yes Yes Yes Severe
3 Yes Yes Yes Yes Yes Yes Yes Yes Severe
4 Yes Yes Yes Yes Yes Yes Yes Yes Severe
5 Yes No Yes No No Yes Yes Yes Modera…
6 No No Yes No Yes Yes Yes Yes Modera…
7 No No Yes No No Yes Yes No Mild
8 No Yes Yes Yes Yes Yes Yes Yes Severe
9 Yes Yes Yes Yes No Yes Yes Yes Modera…
10 No Yes No Yes No Yes No Yes Modera…
# … with 720 more rows, 17 more variables: CoughIntensity <fct>, Myalgia <fct>,
# RunnyNose <fct>, AbPain <fct>, ChestPain <fct>, Diarrhea <fct>,
# EyePn <fct>, Insomnia <fct>, ItchyEye <fct>, Nausea <fct>, EarPn <fct>,
# Pharyngitis <fct>, Breathless <fct>, ToothPn <fct>, Vomit <fct>,
# Wheeze <fct>, BodyTemp <dbl>, and abbreviated variable names
# ¹SwollenLymphNodes, ²ChestCongestion, ³ChillsSweats, ⁴NasalCongestion,
# ⁵SubjectiveFever, ⁶Headache, ⁷Weakness
'data.frame': 730 obs. of 26 variables:
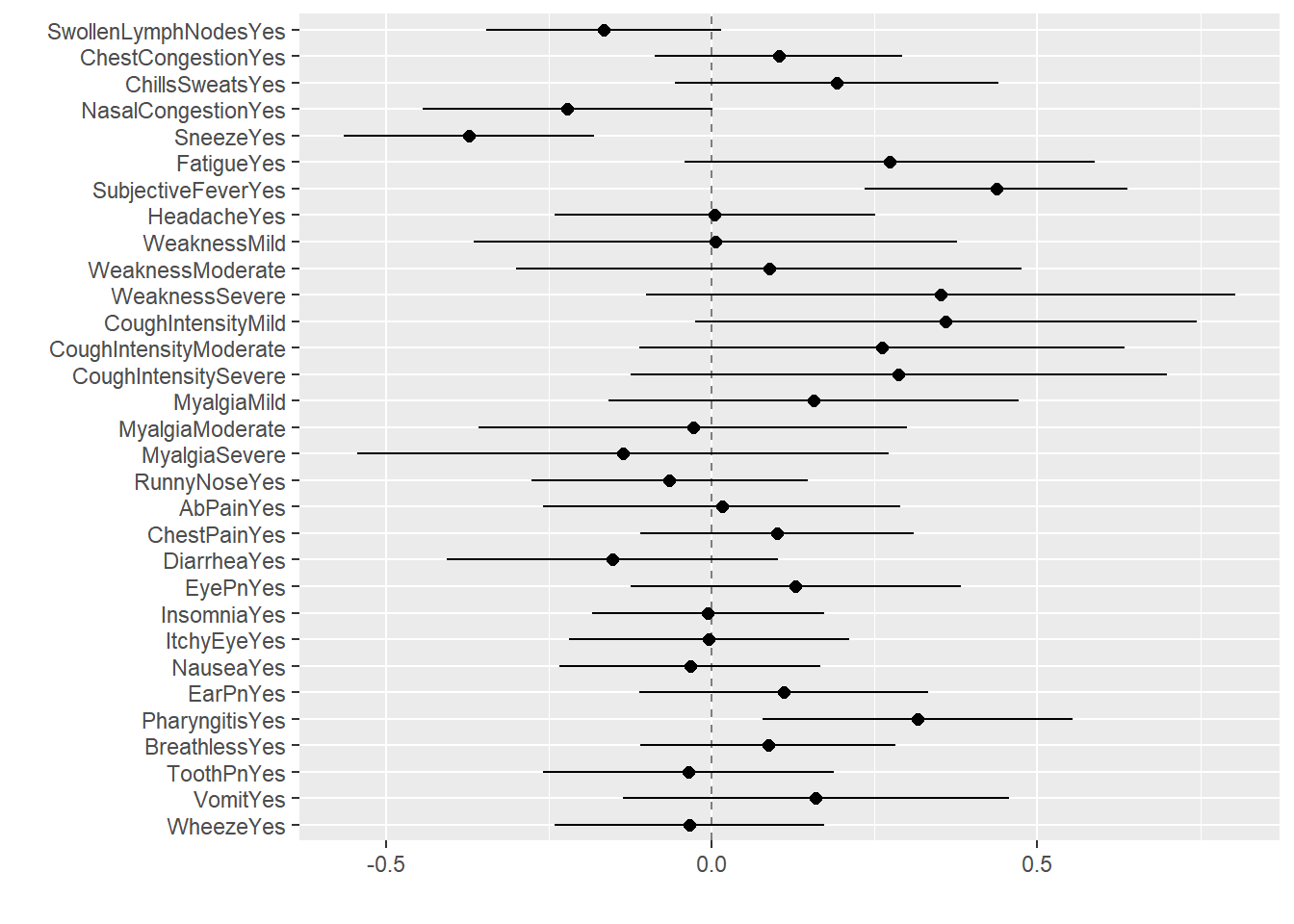
$ SwollenLymphNodes: Factor w/ 2 levels "No","Yes": 2 2 2 2 2 1 1 1 2 1 ...
..- attr(*, "label")= chr "Swollen Lymph Nodes"
$ ChestCongestion : Factor w/ 2 levels "No","Yes": 1 2 2 2 1 1 1 2 2 2 ...
..- attr(*, "label")= chr "Chest Congestion"
$ ChillsSweats : Factor w/ 2 levels "No","Yes": 1 1 2 2 2 2 2 2 2 1 ...
..- attr(*, "label")= chr "Chills/Sweats"
$ NasalCongestion : Factor w/ 2 levels "No","Yes": 1 2 2 2 1 1 1 2 2 2 ...
..- attr(*, "label")= chr "Nasal Congestion"
$ Sneeze : Factor w/ 2 levels "No","Yes": 1 1 2 2 1 2 1 2 1 1 ...
..- attr(*, "label")= chr "Sneeze"
$ Fatigue : Factor w/ 2 levels "No","Yes": 2 2 2 2 2 2 2 2 2 2 ...
..- attr(*, "label")= chr "Fatigue"
$ SubjectiveFever : Factor w/ 2 levels "No","Yes": 2 2 2 2 2 2 2 2 2 1 ...
..- attr(*, "label")= chr "Subjective Fever"
$ Headache : Factor w/ 2 levels "No","Yes": 2 2 2 2 2 2 1 2 2 2 ...
..- attr(*, "label")= chr "Headache"
$ Weakness : Factor w/ 4 levels "None","Mild",..: 2 4 4 4 3 3 2 4 3 3 ...
$ CoughIntensity : Factor w/ 4 levels "None","Mild",..: 4 4 2 3 1 3 4 3 3 3 ...
..- attr(*, "label")= chr "Cough Severity"
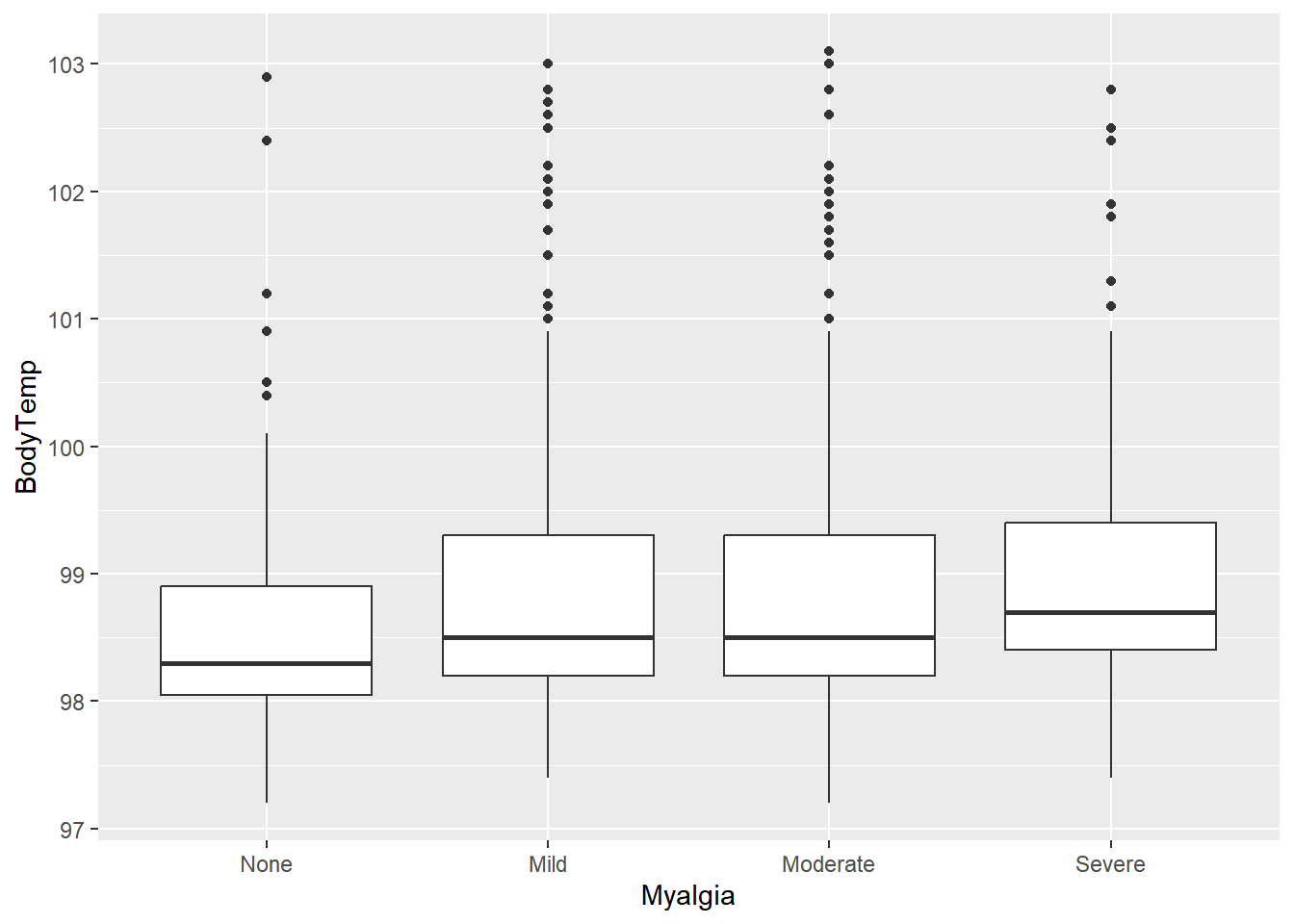
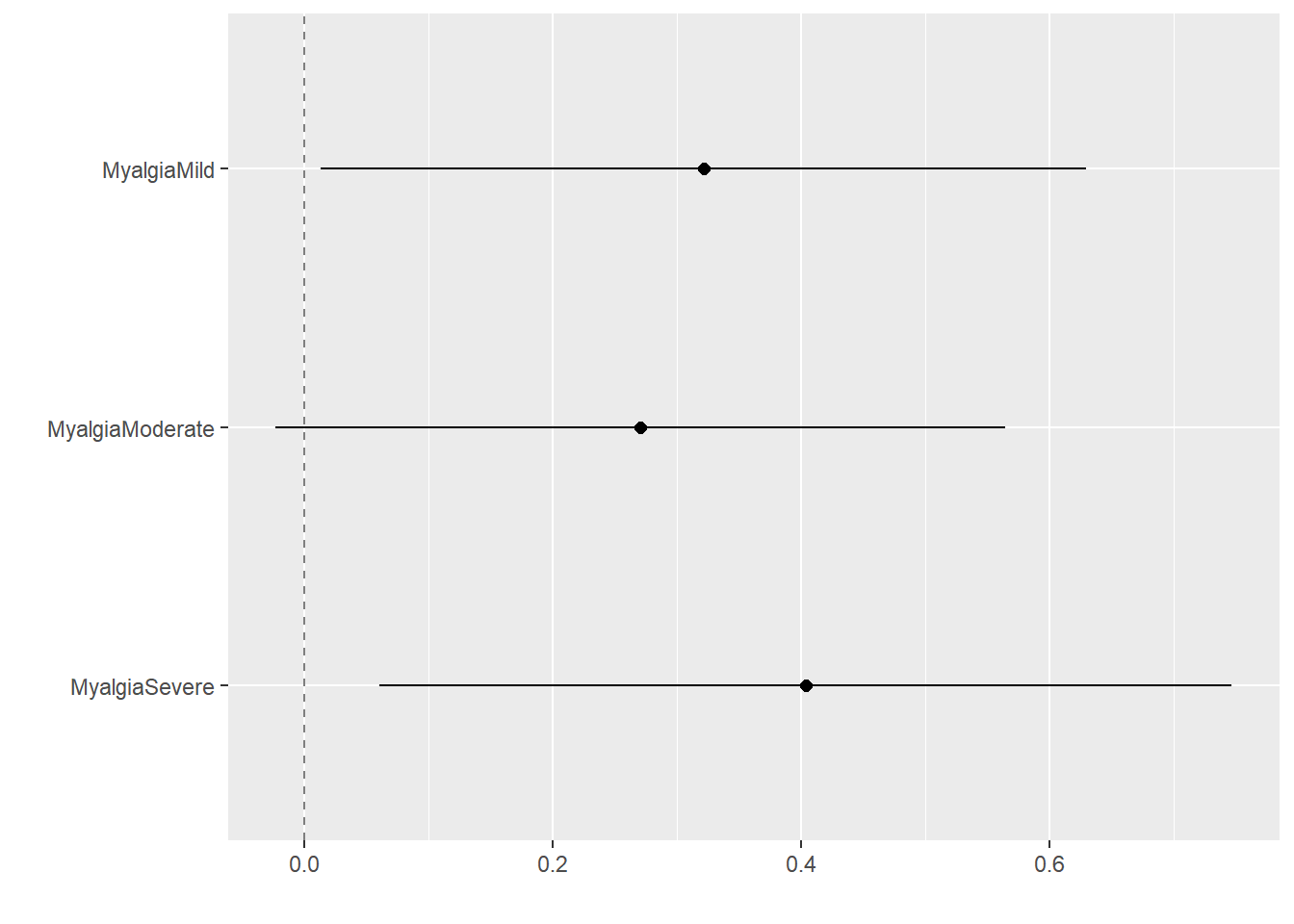
$ Myalgia : Factor w/ 4 levels "None","Mild",..: 2 4 4 4 2 3 2 4 3 2 ...
$ RunnyNose : Factor w/ 2 levels "No","Yes": 1 1 2 2 1 1 2 2 2 2 ...
..- attr(*, "label")= chr "Runny Nose"
$ AbPain : Factor w/ 2 levels "No","Yes": 1 1 2 1 1 1 1 1 1 1 ...
..- attr(*, "label")= chr "Abdominal Pain"
$ ChestPain : Factor w/ 2 levels "No","Yes": 1 1 2 1 1 2 2 1 1 1 ...
..- attr(*, "label")= chr "Chest Pain"
$ Diarrhea : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 2 1 1 1 1 ...
$ EyePn : Factor w/ 2 levels "No","Yes": 1 1 1 1 2 1 1 1 1 1 ...
..- attr(*, "label")= chr "Eye Pain"
$ Insomnia : Factor w/ 2 levels "No","Yes": 1 1 2 2 2 1 1 2 2 2 ...
..- attr(*, "label")= chr "Sleeplessness"
$ ItchyEye : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 1 1 1 1 ...
..- attr(*, "label")= chr "Itchy Eyes"
$ Nausea : Factor w/ 2 levels "No","Yes": 1 1 2 2 2 2 1 1 2 2 ...
$ EarPn : Factor w/ 2 levels "No","Yes": 1 2 1 2 1 1 1 1 1 1 ...
..- attr(*, "label")= chr "Ear Pain"
$ Pharyngitis : Factor w/ 2 levels "No","Yes": 2 2 2 2 2 2 2 1 1 1 ...
..- attr(*, "label")= chr "Sore Throat"
$ Breathless : Factor w/ 2 levels "No","Yes": 1 1 2 1 1 2 1 1 1 2 ...
..- attr(*, "label")= chr "Breathlessness"
$ ToothPn : Factor w/ 2 levels "No","Yes": 1 1 2 1 1 1 1 1 2 1 ...
..- attr(*, "label")= chr "Tooth Pain"
$ Vomit : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 2 1 1 1 ...
..- attr(*, "label")= chr "Vomiting"
$ Wheeze : Factor w/ 2 levels "No","Yes": 1 1 1 2 1 2 1 1 1 1 ...
..- attr(*, "label")= chr "Wheezing"
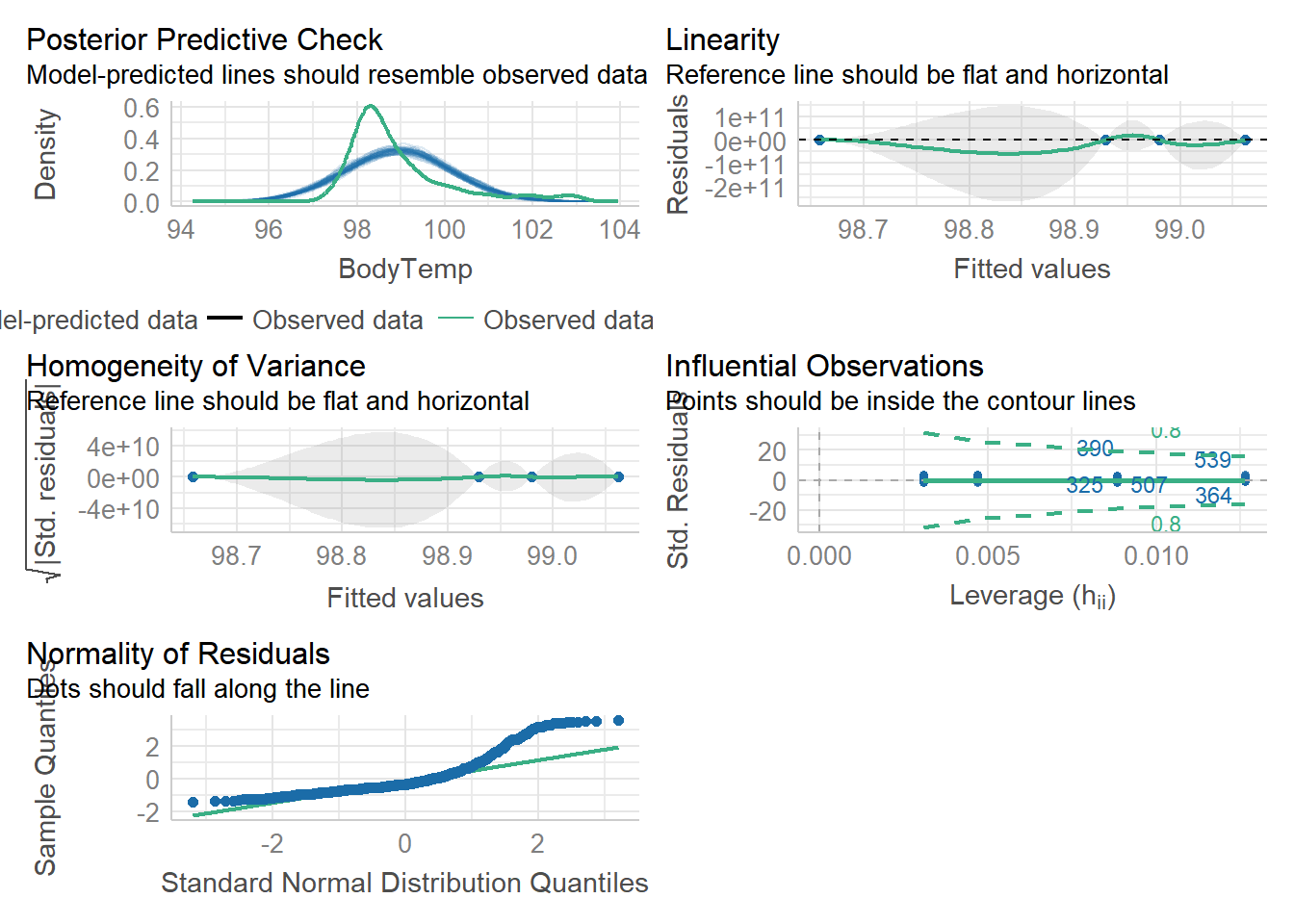
$ BodyTemp : num 98.3 100.4 100.8 98.8 100.5 ...